MEGASAT: High throughput automated genotyping of microsatellites by sequencing.
Zhan L, Paterson IG, Fraser B, Watson B, Bradbury IR, Ravindran PN, Reznick D, Beiko RG, Bentzen P. 2017. MEGASAT: automated inference of microsatellite genotypes from sequence data. Molecular Ecology Resources 17(2):247-256. doi: 10.1111/1755-0998.12561.
Massively parallel DNA sequencing technologies, also known as next generation DNA sequencing (NGS) have revolutionized how genotyping is done in molecular ecology. Most of the newer methods of genotyping target single nucleotide polymorphisms (SNPs), often by sequencing numerous small portions of the genome near restriction enzyme sites (genotyping by sequencing, GBS; restriction site associated markers, RAD tag sequencing or RADseq). However, NGS-based methods also hold the potential to greatly accelerate genotyping of another class of genetic markers, microsatellites.
Microsatellites are short stretches of DNA composed of 2-5 base pair sequences repeated in tandem arrays (e.g., GAGAGAGAGA…) that are embedded in non-repetitive DNA sequences. Organisms typically have hundreds of thousands of microsatellite arrays (‘loci’) scattered around their genomes. The tandem arrays in microsatellites experience slippage mutations that occur at greater rates than mutations elsewhere in the genome; consequently, the number of copies of the repeating unit (‘GA’ in the above example) in a given microsatellite locus often varies among individuals as well as between the two allelic copies of the same microsatellite locus in each (diploid) individual. Because of their abundance in genomes, their high levels of variability (a typical microsatellite may have 10 or more alleles), and their relative ease of assay, microsatellites were the most widely used genetic markers in molecular ecology for the two decades following the early 1990s. However, they have recently been supplanted by SNPs in many applications. One reason for this shift is that the traditional method for genotyping microsatellites, which entails polymerase chain reaction (PCR) amplification of microsatellite loci followed by electrophoretic separation of alleles using denaturing polyacrylamide gels is much less amenable to high throughput genotyping than NGS-based methods that target SNPs (Figure 1).
We developed a high throughput, NGS-based method of genotyping microsatellites that greatly increases genotyping throughput, reduces labour and consumables costs, and maintains a high level of accuracy in genotype calling.
Our method comprises new laboratory protocols, and new software that automates genotype calling. Microsatellites are amplified in multiplex PCRs of up to ~50 loci per PCR, using primers that have 5’ tails containing Illumina adapters (Figure 2). The products of the 1 st round of PCR are then amplified again, in a 2 nd PCR, that adds unique identifier sequences (‘indices’, or ‘barcodes’) to the forward and reverse adapters (Figure 3). By using unique combinations of indices (barcodes) on the forward and reverse adapters, it is possible to uniquely label many individuals. For example, 32 forward adapter indices and 32 reverse adapter indices give 1024 combinations, allowing more than 1,000 individuals to be genotyped in a single sequencing run.
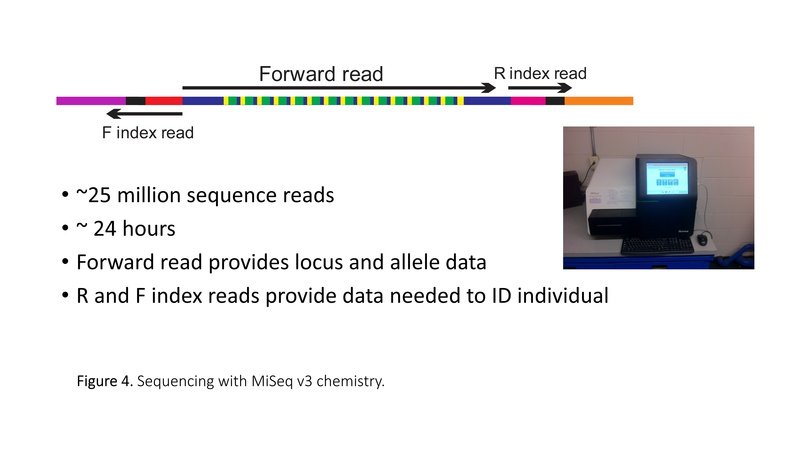
Pooled microsatellite amplicons are sequenced on an Illumina DNA sequencer. Sequencing reads cover the forward and reverse adapter indices (barcodes) and the microsatellite array (Figure 4). Illumina software demultiplexes the sequences, sorting all of the sequences according to the unique combinations of forward and reverse barcodes (Figure 5). MEGASAT sorts the sequences by locus, using primer sequences to identify the locus, and discards any sequences that do not contain a microsatellite (Figure 6). MEGASAT uses decision rules to call the genotypes, and creates graphic output to allow for visualization of data and curation of genotype scores (Figure 7).
Using an Illumina MiSeq DNA sequencer, a single research can genotype many thousands of single-locus genotypes per week, with minimal hands-on labour, a high level of accuracy, and low materials costs (~$0.05-$0.10 CAD per single-locus genotype).